Introduction
Ontology learning[2] is the process of building ontologies automatically from text. Several ontology learning systems [3,1] such as Text2Onto, Doodle OWL and DL-Learner have been developed. Most of these systems support learning of classes, subclasses and taxonomic relationships. But they do not support mining of more expressive axioms from text such as union, intersection, quantifiers and cardinality relation among the concepts. A richer and more expressive ontology can be very useful to the downstream applications such as recommendation systems and question and answering systems.
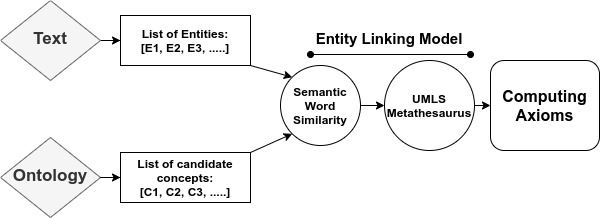
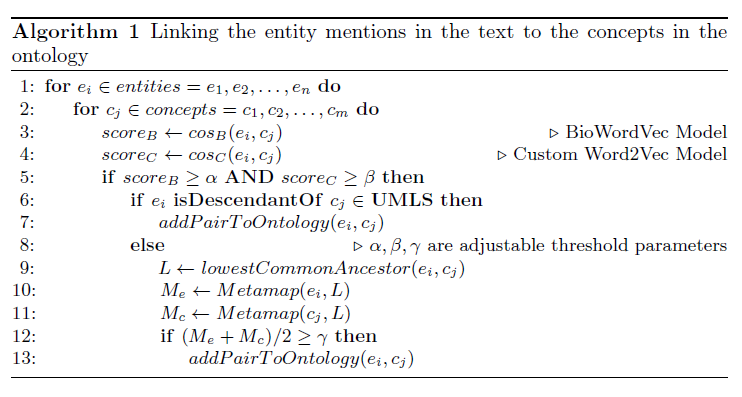
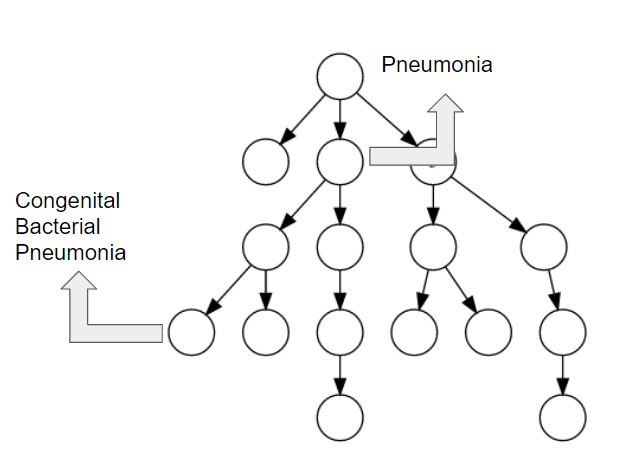
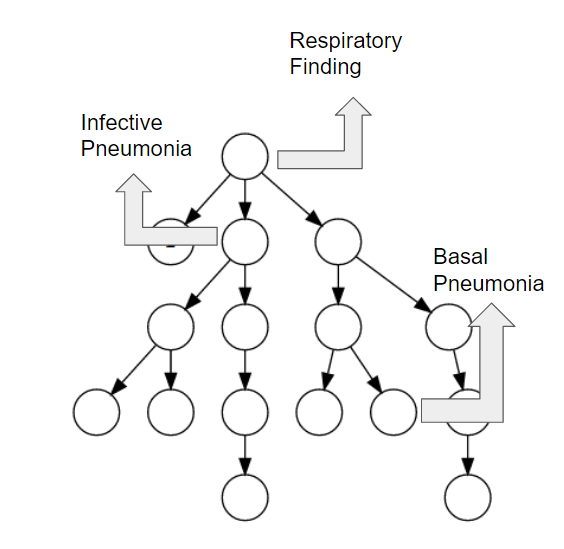
We propose a mechanism to extract union and intersection axioms from the text with the help of an ontology and its relevant text. Examples of intersection and union axioms are given in Axioms 1 and 2. Axiom 1 models the information that a Mixed Glioma (type of tumor) is a combination of Astrocytoma and Oligodendroglioma. Axiom 2 captures the information that a Tumor can be either Benign or PreMalignant or Malignant. Some examples of intersection and union axioms are as below:
Take Relation (1). It can be critical in research of missing properties of any disease that is found as a type of Mixed Glioma based on the other classes of diseases in the relation. That is why, enriching the ontologies with such axioms increase their richness in terms of the information they can provide.